from IPython.core.display import HTML, Markdown, display
import numpy.random as npr
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.stats as stats
import statsmodels.formula.api as smf
import ipywidgets as widgets
import os
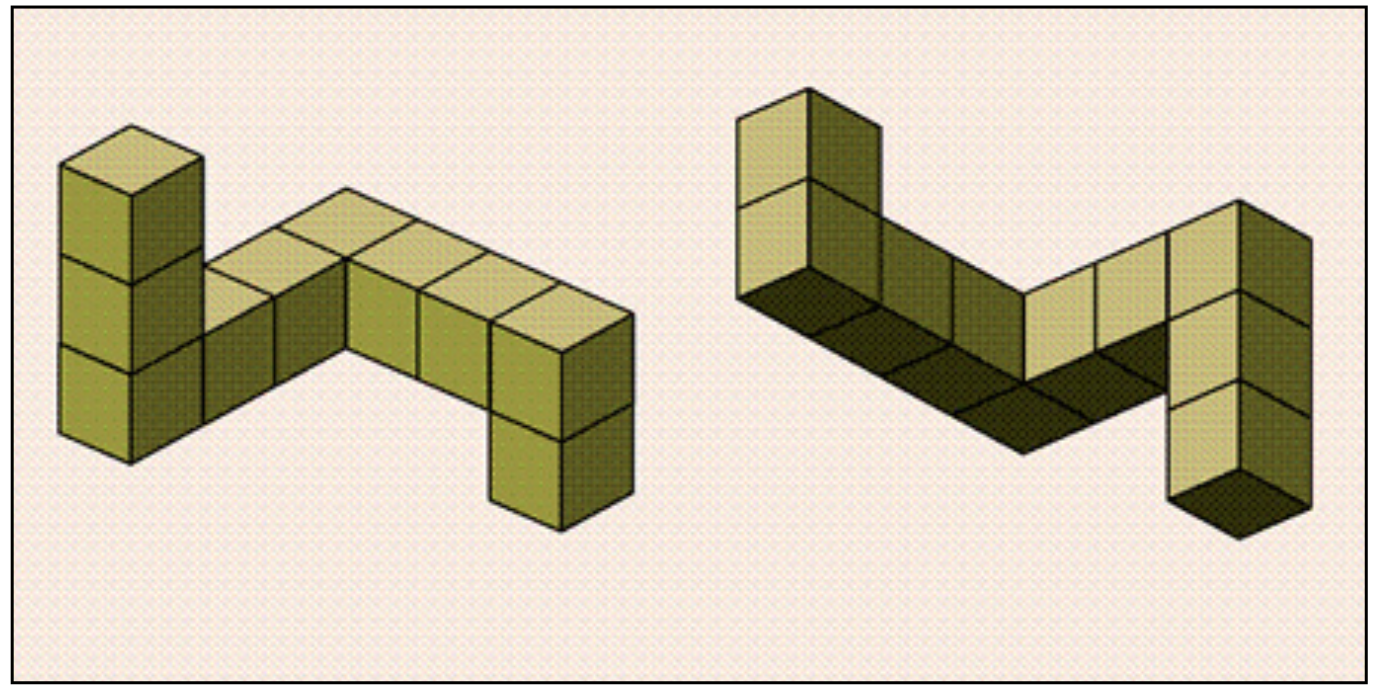
Lab 1C: Mental Rotation¶
As described in the reading, in the Shepard & Metzler (1971) experiments, the time it took people to decide if an object was the same (just rotated) or different (a mirror reflection) depended on the angle of rotation:
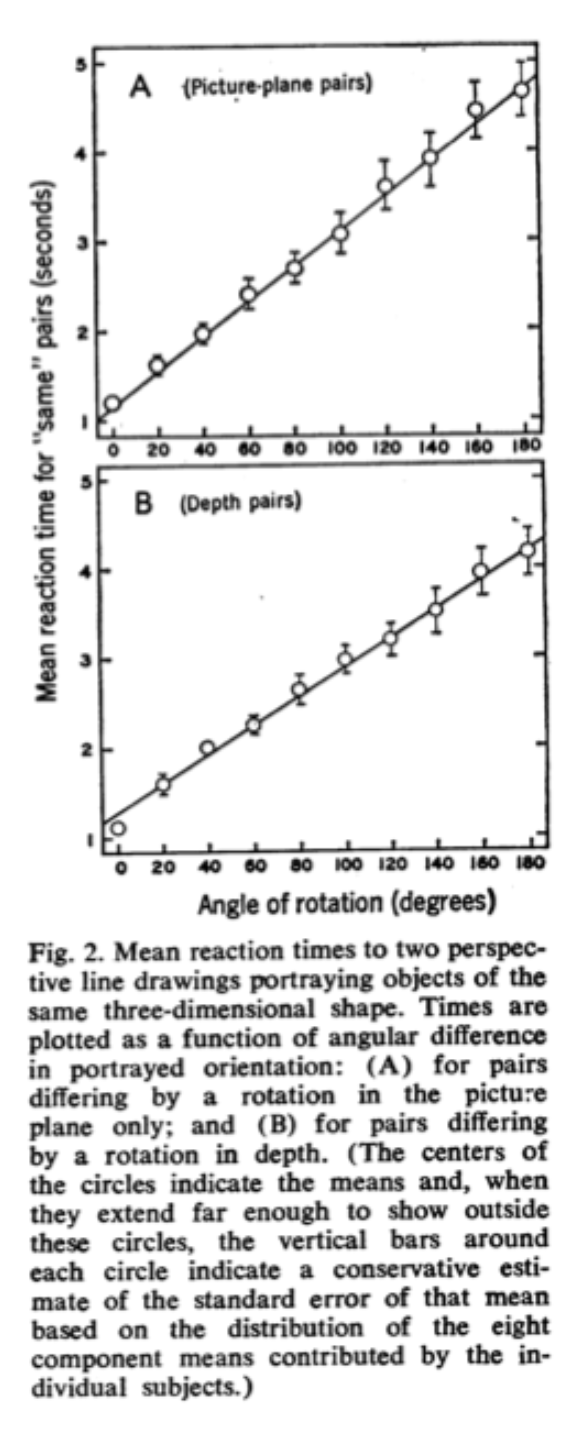
In other words, it would take you longer to decide if the 30 degree rotated version of your cup was the same as the upright version relative to the 15 degree rotated version. The idea is that in order to make the same/mirror judgement, people actually mentally rotate the object. Bigger angles require more time to “rotate” in your mind just as they would in the real world (see Figure above).
Rather than collecting data in class, we will use raw data from this replication of the Shepard and Metzler study (see here). This study ran N=54 participants. Using a for loop, read each of the files from the list below into a pandas dataframe. Then, using the pandas `concat` function (Refer back to the dataframes lab) combine them into a single data frame.
Here is a key to the structure of the dataframe:
Trial: trial number.
Condition: Filename of stimulus
Time: Response time
correct: Correct (desired) response, in other words, the trial type
response: Actual response
angle: Angle (angular disparity, see paper)
correct/incorrect: Was the response correct or incorrect?
# this is an example list comprehension which reads in the all the files.
# the f.startswith() part just gets rid of any junk files in that folder
filenames=['lab2-data/'+f for f in os.listdir('lab2-data') if not f.startswith('.')]
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
Jump to the methods section of the paper where the data comes from, this paper. Read the methods to determine what quality control measures they applied to their data. For instance, what kind of trials were analyzed and which were excluded? Was there any basis for excluding an entire participant? In a markdown cell, describe the set of exclusion criterion that the authors used. Next, create a new data frame of the data from our experiment applying the same exclusion criterion.
Hint: For calculating mean and sd for different groups of trials, I suggest making a new column of the dataframe called ‘tag’ that contains the concatenated strings/fields that define a group. Then you can use the groupby function to find the mean and sd. There are other ways to do this, however.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
Use the pandas 'groupby' function to compute the average reaction time as a function of the angle of the stimulus. It may help to go back to the notebook on dataframes and plotting and searching for the groupby example.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
Using the seaborn 'scatterplot()' function (docs), create a plot of angle versus reaction time collapsing across all the subjects in the task. You should restrict you analysis the same trials that were analyzed in the target reading.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
As you have seen the regplot() and scatterplot() types in seaborn are quite related. Replot the data from above this time using regplot so you can get a sense of the best fit regression line fit to the overall dataset. Again, you should restrict you analysis the same trials that were analyzed in the target reading.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
Using the statsmodels ordinary least squares regression peform a regression of angle (predictor) onto reaction time (outcome) for the whole data frame. Compare the cofficients you obtained to the ones reported in the results section of the paper. (I wouldn't expect the coefficients to match exactly.. they didn't for me)
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
Although in the previous step we might see evidence of the increase of reaction time with angle, there are some caveats because we averaged across a number of other relevant aspects of our data frame. For instance, we averaged across both the subjects (Subject column) and the type of stimuli (if it was 'same' or 'different' - this is in the Correct column). Using the example groupby command displayed in class, find the average reaction time as a function of angle and same/different for each individual subject.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
Using the dataframe you constructed in the last step recreate Figure 3 from the paper. Hint: the plot in Figure three is akin to the 'catplot' in seaborn. Consult the seaborn documentation for information on this plot type.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
The analyses we have considered so far combine across subjects. However we would often like to analyze the effect in terms of individuals. Do individuals show this pattern, or only the group? Let's approach this two ways. One, graphically by making a plot of every individual subject. Two, analytically by running a separate regression model for individual subjects. For the plot use the seaborn `regplot`.
Please conduct this analysis for 3 randomly chosen individual participants.
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.
In your own words, please summarize the results of the analyses you conducted above. Specifically you should mention what the regresssion results imply about the data. How consistent is the pattern across subjects?
Delete this text and put your answer here. The text/code for your analysis should appear in the cells below.